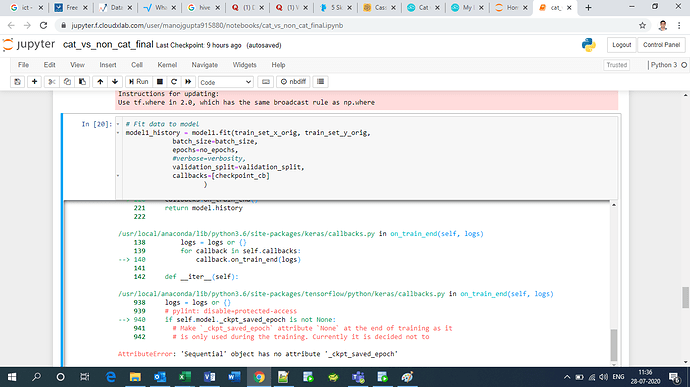
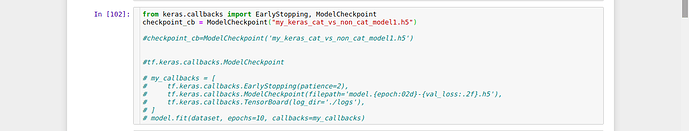
Why callback gives error? I’m not able to understand what is wrong with my code. Can someone please help.
Can I use save_best_only=True when I’m using validation_split=0.2 but not using validation data set separately?
import os as os
import numpy as np
import matplotlib.pyplot as plt
import h5py
import tensorflow
from tensorflow import keras
from PIL import Image
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D
from keras.layers import Activation, Dropout, Flatten, Dense
from keras import backend as K
import numpy as np
from keras.preprocessing import image
# Model configuration
from tensorflow.keras.losses import binary_crossentropy
from tensorflow.keras.optimizers import Adam
seed_value = 42
batch_size = 50
img_width, img_height, img_num_channels = 64, 64, 3 #Image Dimentions of our images. i.e. pixel size and Colour
loss_function = binary_crossentropy
no_classes = 1
no_epochs = 25
optimizer = tensorflow.keras.optimizers.RMSprop() #Adam() #rmsprop
validation_split = 0.2
verbosity = 1
from keras.callbacks import EarlyStopping, ModelCheckpoint
checkpoint_cb = keras.callbacks.ModelCheckpoint("my_keras_cat_vs_non_cat_model1.h5" )
#if we use a validation set during training, you can set save_best_only=True
#checkpoint_cb = keras.callbacks.ModelCheckpoint("my_keras_cat_vs_non_cat_model1.h5", save_best_only=True )
def load_data( path = '/cxldata/datasets/project/cat-non-cat' ):
train_data_path = os.path.join( path, 'train_catvnoncat.h5' )
# Load training data and segregate Features from Labels.
train_data = h5py.File( train_data_path, "r" )
train_data_x = train_data["train_set_x"][:] # train set features (Converting image to numpy array)
train_data_y = train_data["train_set_y"][:] # train set labels
test_data_path = os.path.join( path, 'test_catvnoncat.h5' )
# Load test data and segregate Features from Labels.
test_data = h5py.File( test_data_path, "r" )
test_data_x = test_data["test_set_x"][:] # test set features (Converting image to numpy array)
test_data_y = test_data["test_set_y"][:] # test set labels
classes = np.array( test_data["list_classes"][:] )
return( train_data_x, train_data_y, test_data_x, test_data_y, classes )
train_set_x_orig, train_set_y_orig, test_set_x_orig, test_set_y_orig, classes = load_data( )
# Explore your dataset
print ("Number of training examples: " + str( train_set_x_orig.shape[0] ) ) #train_set_y_orig.size Data size of training data. i.e. no. of images in training set.
print ("Number of testing examples: " + str( test_set_x_orig.shape[0] ) ) #test_set_y_orig.size Data size of validation data. i.e. no. of images in validation set.
print ("Each image is of size: " + str(train_set_x_orig.shape[1:]) )
print ("train_set_x_orig shape: " + str(train_set_x_orig.shape))
print ("train_set_y_orig shape: " + str(train_set_y_orig.shape))
print ("test_set_x_orig shape: " + str(test_set_x_orig.shape))
print ("test_set_y_orig shape: " + str(test_set_y_orig.shape))
#Augmentation for training set.
train_datagen = ImageDataGenerator(
rescale = 1./255,
shear_range = 2.2,
zoom_range = 0.2,
horizontal_flip=True)
#Augmentation for validation set. We will be using less augmentation for validation as we want our validation data to look more natural.
validation_datagen = ImageDataGenerator( rescale=1./255 )
train_datagen.fit( train_set_x_orig )
train_datagen.fit( test_set_x_orig )
# Determine shape of the data
input_shape = (img_width, img_height, img_num_channels)
def define_model1():
model = Sequential()
model.add( Conv2D( 32, (3,3), input_shape=input_shape) )
model.add( Activation( 'relu' ) )
model.add( MaxPooling2D( pool_size = (2, 2) ) )
model.add( Conv2D( 32, (3,3) ) )
model.add( Activation( 'relu' ) )
model.add( MaxPooling2D( pool_size = (2, 2) ) )
model.add( Conv2D( 64, (3,3) ) )
model.add( Activation( 'relu' ) )
model.add( MaxPooling2D( pool_size = (2, 2) ) )
model.add( Flatten() )
model.add( Dense(64) )
model.add( Activation( 'relu' ) )
model.add( Dropout( 0.5 ) )
model.add( Dense( 1 ) )
model.add( Activation( 'sigmoid' ) )
return( model )
def define_model2():
# Create the model
model = Sequential()
model.add( Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape=input_shape))
model.add( Conv2D(64, kernel_size=(3, 3), activation='relu') )
model.add( Conv2D(128, kernel_size=(3, 3), activation='relu') )
model.add( Flatten() )
model.add( Dense(64, activation='relu') )
model.add( Dense(no_classes, activation='sigmoid'))
return( model )
model1 = define_model1()
model2 = define_model2()
model1.summary()
model2.summary()
#from keras import backend as K
#K.clear_session()
model1.compile( loss=loss_function, optimizer='rmsprop', metrics=['accuracy'] )
# Fit data to model
model1_history = model1.fit(train_set_x_orig, train_set_y_orig,
batch_size=batch_size,
epochs=no_epochs,
#verbose=verbosity,
validation_split=validation_split,
callbacks=[checkpoint_cb]
)
model2.compile( loss=loss_function, optimizer='rmsprop', metrics=['accuracy'] )
# Fit data to model
model2_history = model2.fit(train_set_x_orig, train_set_y_orig,
batch_size=batch_size,
epochs=no_epochs,
#verbose=verbosity,
validation_split=validation_split
#callbacks=[checkpoint_cb]
)